
DNA microarrays can be easily interrogated with only the naked eye using a new electrostatic imaging technique developed in the laboratory of Jay Groves, a chemist with Berkeley Lab, UC Berkeley and HHMI. Credit: Photo Michael Barnes, UC Berkeley Chemistry Department The dream of personalized medicine — in which diagnostics, risk predictions and treatment decisions are based on a patient's genetic profile — may be on the verge of being expanded beyond the wealthiest of nations with state-of-the-art clinics. A team of researchers with the U.S. Department of Energy's Lawrence Berkeley National Laboratory (Berkeley Lab) has invented a technique in which DNA or RNA assays — the key to genetic profiling and disease detection — can be read and evaluated without the need of elaborate chemical labeling or sophisticated instrumentation. Based on electrostatic repulsion — in which objects with the same electrical charge repel one another — the technique is relatively simple and inexpensive to implement, and can be carried out in a matter of minutes.
"One of the most amazing things about our electrostatic detection method is that it requires nothing more than the naked eye to read out results that currently require chemical labeling and confocal laser scanners," said Jay Groves, a chemist with joint appointments at Berkeley Lab's Physical Biosciences Division and the Chemistry Department of the University of California (UC) at Berkeley, who led this research. "We believe this technique could revolutionize the use of DNA microarrays for both research and diagnostics." 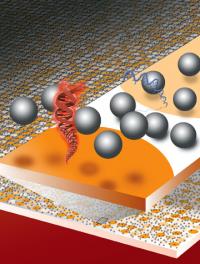
A new method for reading DNA (or RNA) microarrays is based on measuring the electrostatic repulsion between silica microspheres and hybridized DNA. Surface areas containing double-stranded DNA (red) or single-stranded DNA (blue) can be easily distinguished from each other and from background areas by the naked eye. Credit: Illustration by Flavio Robles, Berkeley Lab Public Affairs
Groves, who is also a Howard Hughes Medical Institute (HHMI) investigator, and members of his research group Nathan Clack and Khalid Salaita, have published a paper on their technique in the journal Nature Biotechnology, which is now available online. The paper is entitled "Electrostatic readout of DNA microarrays with charged microspheres."
In their paper, Groves, Clack, and Salaita describe how dispersing a fluid containing thousands of electrically-charged microscopic beads or spheres made of silica (glass) across the surface of a DNA microarray and then observing the Brownian motion of the spheres provides measurements of the electrical charges of the DNA molecules. These measurements can in turn be used to interrogate millions of DNA sequences at a time. What's more, these measurements can be observed and recorded with a simple hand-held imaging device — even a cell phone camera will do.
"The assumption has been that no detection technique could be more sensitive than fluorescent labeling, but this is completely untrue, as our results have plainly demonstrated," said Groves. "We've shown that changes in surface charge density as a result of specific DNA hybridization can be detected and quantified with 50-picometer sensitivity, single base-pair mismatch selectivity, and in the presence of complex backgrounds. Furthermore, our electrostatic detection technique should render DNA and RNA microarrays sufficiently cost effective for broad world-health applications, as well as research."
Your susceptibility to a given disease and how your body will respond to drugs or other interventions is unique to your genetic makeup. Under a personalized medicine plan, treatment effectiveness is maximized and risks are minimized by tailoring disease treatments specifically to you. This requires the precise diagnostic tests and targeted therapies that can stem from assays using a DNA microarray — a thumbnail-sized substrate containing thousands of microscopic spots of oligonucleotides (stretches of DNA about 20 base pairs in length) laid out in a grid.
Often referred to as "gene chips," DNA microarray assays and their RNA counterparts have become one of the most powerful tools for gene-expression profiling, the identification of mutations, and the detection of multiple pathogens in patients afflicted either by multiple diseases or drug-resistant strains of diseases. Aside from their potential future role in personalized medicine, the widespread use of DNA microarray assay devices could have an immediate and profound impact on the treatment of diseases today. For example, according to a report two years ago from the Global Health Diagnostics Forum, 400,000 lives could be saved each year from death by tuberculosis through the use of DNA microarray assays rather than the standard TB diagnostic test, which is known to miss nearly half of all cases.
Until now, however, the use of DNA microarray assays has been limited because current techniques typically depend upon fluorescence detection, a demanding methodology that requires time-consuming chemical labeling, high-power excitation sources, and sophisticated instrumentation for scanning. Such demands are generally well beyond the capabilities of individual laboratories or clinics, especially in developing countries. While label-free DNA detection strategies do exist, they require either complex device fabrication or sophisticated instrumentation for readouts, and in addition none are compatible with conventional DNA microarrays, where up to one million sequences are available for interrogation in a single experiment.
"We have demonstrated parallel sampling of a microarray surface with micron-scale resolutions over centimeter-scale lengths," said Groves. "This is four orders of magnitude larger than what has been achieved to date with conventional scanning-electrostatic-force microscopy."
In a typical experiment, a microarray is prepared and mounted in a well chamber and the DNA is hybridized (a standard technique in which complementary single strands of DNA bind to form double-stranded DNA "hybrids"). A suspension of negatively-charged silica microspheres is then dispersed through gravitational sedimentation over the microarray surface, a process which takes about 20 minutes. Because the substrate or background surface of the microassay is positively charged, the silica microspheres will spread across the entire surface and adhere to it. However, on surface areas containing double-stranded DNA, which is highly negatively charged, and on areas containing single-stranded DNA, also negatively charged but to a lesser degree than double-stranded DNA, the microspheres will levitate above the substrate surface, stacking up in "equilibrium heights" that are dictated by a balance between gravitational and electrostatic forces.
These electrostatic interactions on the microarray surface result in charge-density contrasts that are readily observed. Surface areas containing DNA segments take on a frosted or translucent appearance, and can be correlated to specific hybridizations that reveal the presence of genes, mutations and pathogens.
"Our technique is essentially a millionfold parallel version of the classic experiment used by Robert Millikan almost 100 years ago, when he determined the charge of a single electron by observing the positions of oil droplets levitated above a charged plate," said Groves.
There are a number of short-term "next steps" for this research, Groves said, including testing its application in high-density arrays and pushing its ultimate resolution limits.
"Since the resolution of electrostatic-based imaging is determined by the number of particle-observations rather than by the diffraction limit of light, our readouts could serve as a form of ultramicroscopy," he said. "The real grand challenge for this technology, however, will be for us to find suitable industrial partners with whom we can work to see that useful new products actually make it to market."
Source : DOE/Lawrence Berkeley National Laboratory
 Print Article
Print Article Mail to a Friend
Mail to a Friend
