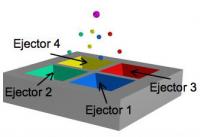
Each reservoir contains a different base or 'letter' in the DNA alphabet. When activated, an acoustic membrane located underneath the membrane aims a tiny droplet of the desired letter to a point above the center of the array, to the surface of a slide (not shown) successive activations build up a DNA sample on the slide. Credit: USC Ming Hsieh, department of electrical engineering A University of Southern California researcher who with his team won a prize last year for demonstrating a new way to create DNA microarrays has improved the technique to make it much more powerful.
DNA microarrays, also known as gene chips, are a basic tool for modern medicine and biology, allowing researchers to detect whether a certain message sequence is present or absent in a given sample.
Usually, these chips are made to order by specialty companies, using extremely expensive equipment. But the directional soundwave technique invented by the research group of Eun Sok Kim, a professor at the USC Viterbi School's Ming Hsieh department of electrical engineering, will potentially allow researchers to have their own gene chip making machines in their own labs.
The Kim group's machines consist of an array of quartets of ejectors, depressions in which small drops of DNA bases -- the genetic chemical letters A, T, C, and G -- sit on top of thin membranes that can be given electronic snaps by an activator. 
Photograph of a packaged ejector array.
The snap is directional and extremely precise, so that droplets emerge at exactly the same predetermined angle from the vertical. The paths from the four different ejectors converge at a single point about 2 millimeters above the quartet package, adhering there to the surface of a slide. For each snap, only one of the four ejectors fires.
Supply lines from four central reservoirs refill the injectors after each snap.
Construction of quartet array system, described as a "nozzleless, heatless, lensless" machine is a complex process Kim and his former students, Jae Wan Kwon, now of the University of Missouri/Columbia and Sanat Kamal-Buhl, now of Intel, first described in an April 2006 paper in the IEEE Transactions on Automation Science and Engineering paper.
That paper won the journal's "Best New Application Paper Award" for 2006, based on the criteria of " technical merit, originality, potential impact on the field, clarity of presentation, practical significance for applications, and the novelty of the new application." The honor carries a cash prize and certificate to be awarded at a conference September 22.
In the paper Kim and his co-authors discussed the advantages of the system over existing techniques, which include photomask and inkjet-like printing systems and and also painstaking contact transfer.
Inkjet systems also eject droplets, but "the smallest droplet size depends on the size of the nozzle, but the small nozzles are difficult to construct with uniformity and tend to get clogged," the paper notes.
Manufacturers in the field include Affymetrix, Agilent, and Rosetta.
In the 2006 paper, the authors said that they were working on a way to synthesize sequences longer than the 3-base n-mers the pilot machine produced.
They have now done so. A new paper accepted for publication in IEEE/ASME Journal of Micro-Electro-Mechanical Systems, details how the system has been improved to allow synthesis of 15-mers. Kim and his co-workers have patents pending on the process.
A working commercial machine is still at least 3-5 years away, says Kim. "It would require better integration of the ejectors, microchannels, and interconnecting ports, miniaturization of the driving electronics and wash/dry station, and packaging of the whole system."
He estimates that the first models would cost in the neighborhood of $100,000.
Source : University of Southern California
 Print Article
Print Article Mail to a Friend
Mail to a Friend
