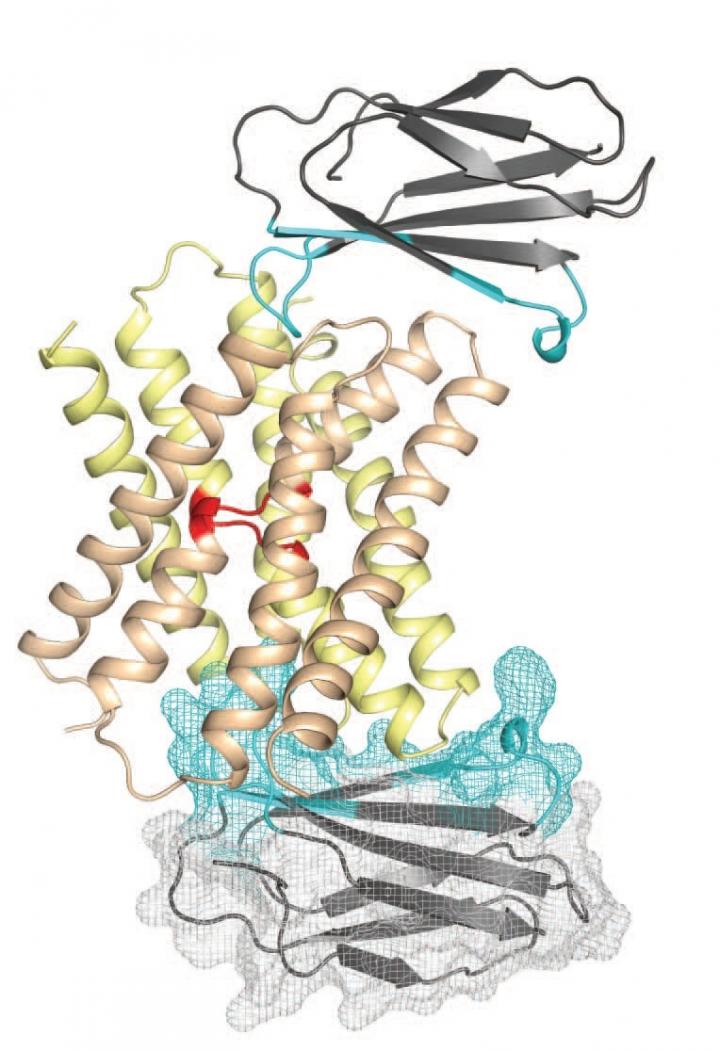
This is a representation of the fluoride ion channel (center) with monobodies attached (top and bottom). Although present almost everywhere - food, soil, toothpaste and especially tap water -, the fluoride ion is highly toxic to microorganisms and cells. To avoid death, cells must remove fluoride that has accumulated inside them, a process accomplished via ion channels - protein tunnels through the cell membrane that only allow specific substances to pass through.
Fluoride ion channels have only recently been discovered, and studies have hinted at an unusual structure that might explain their remarkable selectivity for fluoride. However, these channels are small and have been extremely challenging to crystallize, which has thus far prevented investigations of their atomic structure.
In a study published in Nature on Sept. 7, 2015, an international team of scientists overcame these limitations and for the first time resolved the atomic structure of a fluoride ion channel. They discovered a unique "double-barreled" architecture that contains two pathways through which fluoride ions flow, representing a new mechanism of ion transport. Their findings shed light on the evolution of these channels and enable new approaches to modify their function, with potential applications such as the development of novel antibiotics.
The key technological innovation that enabled this achievement was the use of monobodies - small, synthetic proteins that can be custom-designed to bind to highly specific locations on target proteins. Monobodies were engineered to function as 'crystallization chaperones' that stabilized and aligned fluoride ion channels so their structure could be determined.
"It is very clear that synthetic binding technologies are now mature," said study co-author Shohei Koide, PhD, professor of biochemistry and molecular biophysics at the University of Chicago and a world-leader in monobody research and design. "Monobodies are refined and powerful enough to attack important questions in biology and medicine, such as the structure of the fluoride ion channel, but they can also be used to generate molecules that are almost immediately useful in industry. It's exciting to us as we continue to reveal the many possibilities of this powerful technology."
The Fluc family of fluoride ion channels were recently discovered, and studies by a team led by Christopher Miller, PhD, HHMI Investigator and professor of biochemistry at Brandeis University, found that they are among the most selective channels yet identified. It has remarkable specificity for fluoride and can distinguish it from the closely related chloride ion.
To determine the atomic structure of these channels, a technique known as x-ray crystallography must be used. This requires that the channel protein be purified and crystallized (aligned in a highly ordered, reproducible manner). However, membrane proteins are unstable when removed from their native environment. To circumvent this, traditional biochemical techniques use detergent compounds to mask and stabilize the surface areas that once resided in cell membrane, allowing crystallization to occur.
For the fluoride ion channel, the surface area that is covered by detergent compounds proved to be too much for crystal formation to occur, due to the channel's small size and almost complete embedment within the membrane.
To address this challenge, Koide, Miller and their teams designed monobodies that bound to target locations on the fluoride ion channel - specifically, two small surfaces not embedded within the cell membrane. This dramatically expanded the surface area that could be used to crystallize the channels. In addition, monobodies have certain properties that allow them to align with each other in a highly-reproducible fashion, further enabling the creation of a stable lattice. The two groups further collaborated with Simon Newstead, PhD, and his team from the University of Oxford, and utilized new crystallization technology for membrane proteins. This three-way collaboration yielded the atomic structure of the channel.
The researchers discovered that the fluoride ion channel is comprised of two major building blocks, which assemble together in an antiparallel manner. Most typical ion channels have only one pore through which ions¬¬ flow in the entire channel assembly, but the Fluc channel was found to have two. This architecture resembles another class of membrane proteins called transporters that use energy to actively pump ions across the cell membrane. However, Fluc channels do not require energy. This similarity and distinction led the researchers to propose that the term "channsporter" could be applied to these channels.
"The function of this molecule is a channel, but the architecture is more like a transporter," Koide said. "Evolutionarily, it's quite interesting. It requires further study, but this protein appears as if it evolved from something that probably had antiparallel geometry that was important for integrity, not necessarily function."
The monobodies also represent the first known molecules, natural or synthetic, that block a Fluc channel. The structure with a blocker allowed a deeper analysis of how fluoride ions flow through the channel. While this study has provided clues as to the channel's remarkable sensitivity, additional studies are underway to further elucidate its physical dynamics.
The ability to specifically block the ion channel with monobodies also raises intriguing possibilities for translational applications. For example, blocking the fluoride ion channel in undesirable microorganisms such as pathogenic bacteria would cause a lethal accumulation of fluoride when they are exposed to something as common as tap water. This could represent a completely new strategy for the development of potent antibiotics.
"With the structure now known, we can figure out exactly how fluoride ion channels work and how to control their function," Koide said. "It's blue sky, but we can certainly envision making different inhibitors of channels with monobodies, which could perhaps be targeted against harmful cells and microorganisms."
"For now, this study represents a proof of concept that monobodies can be designed to work with very difficult targets to make inhibitors and determine structural information," he adds. "We are now working with collaborators on similar applications in many other systems."
Source: University of Chicago Medical Center
 Print Article
Print Article Mail to a Friend
Mail to a Friend
