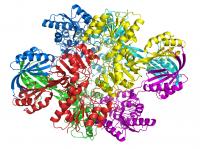
These simplified images from Wayne Anderson's lab show how the polypeptide backbone of a Bacillus anthracis protein folds and curls around itself like a ribbon. It is one of 5,617 proteins in the bacteria that cause anthrax. Credit: Wayne Anderson A scientist slides on a pair of plastic 3-D glasses and an unearthly blue multi-armed creature -- an image right out of a sci-fi horror flick -- seems to leap out of the computer screen into the laboratory.
But this is no movie director's fantasy. The horror image is real.
The eerie "creature" is from the deadly anthrax bacteria -- specifically one of its proteins. Scientists at Northwestern University's Feinberg School of Medicine are mapping parts of the lethal bacteria in three dimensions, exposing a new and intimate chemical portrait of the biological killer down to its very atoms. This view of the disease will offer scientists who design drugs a fresh opening into the bacteria’s vulnerabilities, and thus enable them to create drugs to disable it or vaccines to prevent it.
Anthrax is just the beginning. The Feinberg School is directing an ambitious national project that will map a rogues' gallery of 375 proteins from deadly infectious diseases over the next five years. It is being funded by a $31 million contract from the National Institute of Allergy and Infectious Diseases, part of the National Institutes of Health. The payoff could be a wave of new medicines to wipe out some of the worst scourges to ever infect the human race.
"The concept is fairly simple," said Wayne Anderson, who is leading the national project at the Feinberg School's new Center for Structural Genomics of Infectious Diseases. "If you have a lock and a key and you don't know what either one looks like, how will you design them to fit together"" The lock is Anderson's metaphor for the disease; the key is the drug or vaccine that will slip inside its atomic structure and destroy it.
To figure out where to throw the chemical equivalent of a monkey wrench into the anthrax cell -- and others --Anderson will be mapping key proteins the bacteria uses to do its work.
"We'll see what the proteins look like and see what they need to grow so scientists can use this information to design drugs to knock them out," explained Anderson, a professor of molecular pharmacology and biological chemistry. “We might look at a protein that copies a virus' or bacteria’s genome so it can infect people. If we can find a chemical to stop it from working, it prevents the virus from reproducing and spreading infection.”
The proteins in his lab, by the way, are not capable of triggering an infectious disease. "You need the actual virus or bacteria for that," Anderson said.
Eventually Anderson's gallery will be filled with the not-so-pretty genetic portraits of proteins from the plague, cholera, rabies, West Nile virus, viral encephalitis and Ebola, just to name a few. He'll also be looking at newly emerging diseases and drug resistant infections. His team -- which includes researchers at seven other institutions -- will churn out the three dimensional atomic structures of at least 75 disease proteins a year and quickly post their discoveries on a special website for scientists who can immediately use the information to work on new drugs.
This mega-assault on these diseases at such dizzying speed, scientifically speaking, represents a tectonic shift in how researchers are attacking infectious diseases.
Up until now, molecular pharmacologists -- the people who design new medicines -- had to work at a much slower pace because they had access to only one protein image from a disease at a time.
New state-of-the art technology has accelerated the process. "Now we are going through the genome and finding 100 proteins from a bacteria," explained Anderson. "We're looking at all of these and providing the information so scientists can look at more than one at a time."
To obtain these unusual proteins for their "photo op," Anderson first has to grow them into crystals. A few steps from his office is the “nursery,” where hundreds of thousands of viral and bacterial protein crystals grow in what resemble miniature ice-cube trays. The trays are stacked in giant incubators to keep the proteins at their favorite temperature.
Because Anderson is never sure what environment will produce a crystal -- some proteins prefer more acidity or salt than others, he tries hundreds of different recipes for each one.
Viewing these proteins down to the arrangement of their atoms requires an intense x-ray beam. One of the few sites in the country with this technology is the enormous Synchrotron at Argonne National Laboratory. From the air, the Synchrotron looks like an indoor track. And, in a way, it is. The only runners, however, are electrons circling the Synchrotron, which is actually a 40-sided polygon one kilometer around. As the electrons race around the polygon, they shoot off intense x-ray beams.
Working with equipment inside a special lead-walled station to protect them from radiation, scientists place a protein crystal -- just 1/10 of a millimeter -- into the Synchrotron x-ray beam. As the x-rays scatter off the crystal, the diffraction pattern reveals the location of the protein's electrons and atoms, a process called x-ray crystallography.
In early November, the university will launch a website, www.csgid.org, for scientists who specialize in different bacteria or viruses, so they can scan the project's list of infectious diseases and suggest which proteins Anderson and his colleagues should examine. “We hope we’ll get suggestions from people in the scientific community," said Anderson, who also is co-director of Northwestern's Synchrotron Research Center. "Their knowledge can be a big help to us because each bacteria makes thousands of proteins." The website also will be continuously updated to show scientists newly mapped proteins.
"We hope our effort will lay the groundwork for new drugs to treat or prevent some of the worst infectious diseases to plague our country and the world," Anderson said.
Source : Northwestern University
 Print Article
Print Article Mail to a Friend
Mail to a Friend
