A super-resolution technique developed at Rice University allows fluorescent-labeled probe DNA to pinpoint target DNA sequences in an immobilized strand in ways neither regular nor electron microscopes are able. With high-tech optical tools and sophisticated mathematics, Rice University researchers have found a way to pinpoint the location of specific sequences along single strands of DNA, a technique that could someday help diagnose genetic diseases.
Proof-of-concept experiments in the Rice lab of chemist Christy Landes identified DNA sequences as short as 50 nucleotides at room temperature, a feat she said is impossible with standard microscopes that cannot see targets that small, or electron microscopes that require targets to be in a vacuum or cryogenically frozen.
The technique called "super-localization microscopy" has been known for a while, Landes said, but its application in biosensing is just beginning.
The work by Landes, Rice postdoctoral associate Jixin Chen and undergraduate student Alberto Bremauntz is detailed in the American Chemical Society journal Applied Materials and Interfaces.
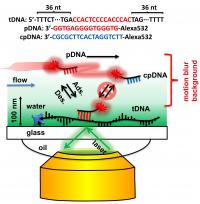
The Rice researchers call their super-resolution technique "motion blur point accumulation for imaging in nanoscale topography" (mbPAINT). With it, they resolved structures as small as 30 nanometers (billionths of a meter) by making, essentially, a movie of fluorescent DNA probes flowing over a known target sequence along an immobilized single strand of DNA. __IMAGE_2
The probes are labeled with a fluorescent dye that lights up only when attached to the target DNA. In the experimental setup, most would flow by unseen, but some would bind to the target for a few milliseconds, just long enough to be captured by the camera before the moving liquid pulled them away. Processing images of these brief events amidst the background blur allows the researchers to image objects smaller than the natural diffraction limits of light-based imaging, which do not allow for the resolution of targets smaller than the wavelength of light used to illuminate them.
Even the Landes lab's system is subject to these physical limitations. Individual images of fluorescing probes on targets are just a pixelated blur. But it's a blur with a bright spot, and careful analysis of multiple images allows the researchers to pinpoint that spot along the strand.
"The probes are moving so fast that in real time, all we would see with the camera is a line," Chen said. But when the camera firing at 30-millisecond intervals happened to catch a bound probe, it clearly stood out. The probes sometime picked out two sequences along a strand that would have been seen as a single blur via regular fluorescent microscopy.
Landes said one goal for mbPAINT is to map ever-smaller fragments of DNA. "Eventually, we'd like to get down to a couple of nucleotides," she said. "Some diseases are characterized by one amino acid mutation, which is three nucleotides, and there are many diseases associated with very small genetic mutations that we'd like to be able to identify.
"We're thinking this method will be ideally suited for diseases associated with small, localized mutations that are not possible to detect in any other inexpensive way," she said.
Landes sees mpPAINT as not only more cost-effective but also able to capture information electron microscopes cannot.
"One of the reasons people invented electron microscopy is to image objects smaller than light's diffraction limit, because biomolecules such as proteins and DNAs are smaller than that," she said. "But electron microscopy requires cryogenic temperatures or a vacuum. You can't easily watch things react in solution.
"The advent of this technology allows us to see the biological processes of nano-sized objects as they happen in water, with buffers and salts, at room temperature, at body temperature or even in a cell. It's very exciting," Landes said.
Source : david@rice.edu
 Print Article
Print Article Mail to a Friend
Mail to a Friend
